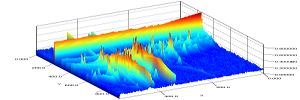
PairwiseAlignmentMatrixVisualizer
Class: maltcms.commands.fragments.visualization.PairwiseAlignmentMatrixVisualizer
Workflow Slot: VISUALIZATION
Description: Creates a plot of the pairwise distance and alignment matrices created during alignment.
Variables
Required
var.total_intensity var.pairwise_distance_matrix var.pairwise_distance_alignment_names
Optional
var.anchors.retention_index_names var.anchors.retention_scans
Provided
Configurable Properties
Name: chromatogramHeight
Default Value: 200
Description:
The height of the chromatogram profile in pixels.
Name: colorramp_location
Default Value: res/colorRamps/bw.csv
Description:
The location of the color ramp used for plotting.
Name: drawPath
Default Value: true
Description:
If true, plot the alignment path.
Name: fontsize
Default Value: 30
Description:
The font size used for labels in pt.
Name: format
Default Value: png
Description:
The file format to save plots in.
Name: full_spec
Default Value: false
Description:
Deprecated
Name: left_chromatogram_var
Default Value: total_intensity
Description:
Name: matrix_vars
Default Value: [cumulative_distance, pairwise_distance]
Description:
A list of matrix variables to plot
Name: pairsWithFirstElement
Default Value: false
Description:
If true, plot only pairs with first chromatogram.
Name: path_i
Default Value: null
Description:
Name: path_j
Default Value: null
Description:
Name: sampleSize
Default Value: 1024
Description:
The number of color samples used in plots.
Name: top_chromatogram_var
Default Value: total_intensity
Description: